BioSET: Biomarker-based Spatial co-Expression analysis in Tumor environments
Award of Excellence
Chahat Kalsi, Yuancheng Shen, Sophia Gaupp, Luca Reichmann, Meri Rogava, Michael Krone, Saeed Boorboor, Robert Krüger
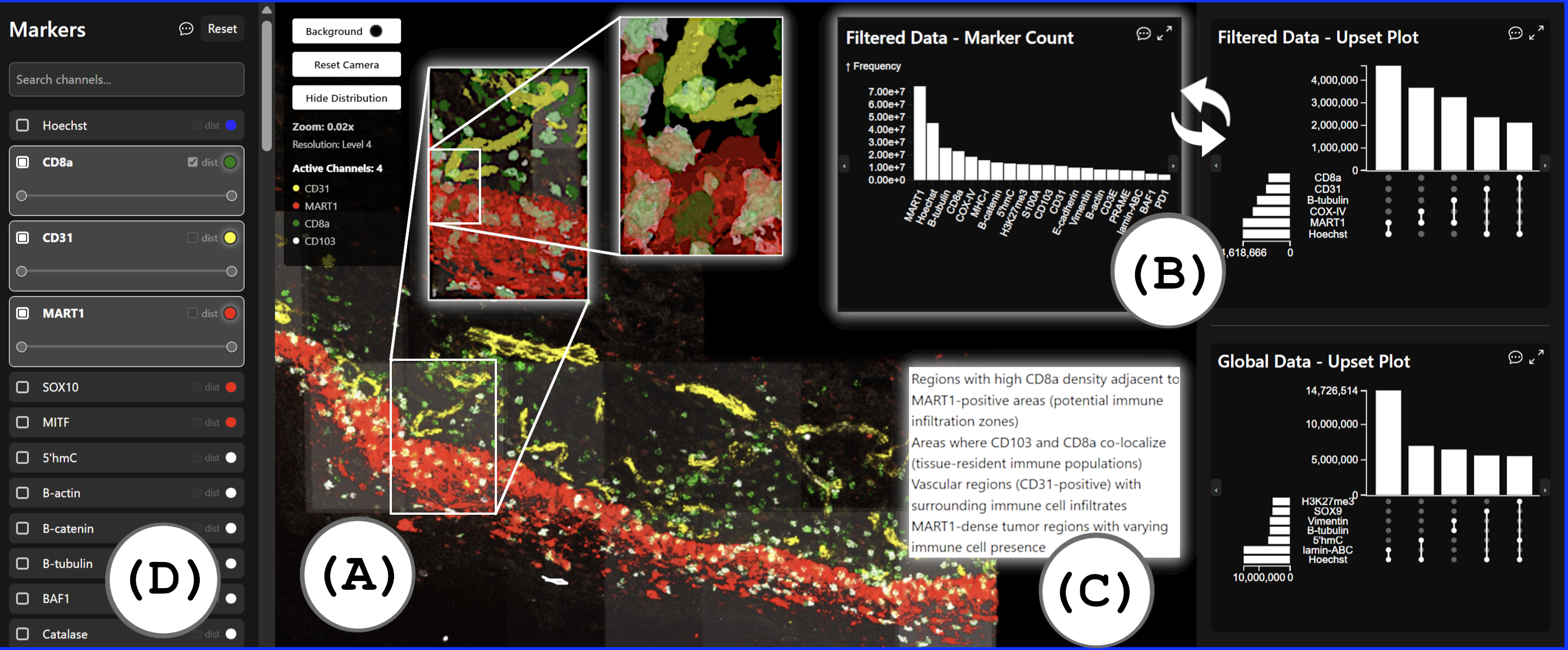
This abstract reports on our solution to the IEEE VIS 2025 Bio+MedVis 3D Microscopy Imaging Challenge with the tasks of identification and visualization of cell-cell interactions in 3D Cyclic Immunoflorescence (CyCIF) data. The provided 3D melanoma tissue data comprises 70 channels (volumes) across 6 levels of resolution, the highest having 194x5,508x10,908 voxels per channel (1.48,TB) and the lowest having 194x172x340 voxels per channel (1.48,GB). Our approach makes the following contributions: (1) a method to detect protein-expressing regions and identify spatially co-located marker sets in the tissue. (2) A scalable overview-detail visualization, combining multi-scale volume and surface rendering, to guide users to regions of interest, and (3) integrated contextual querying, providing semantic explanations of displayed patterns. We analyzed the data using our tool in joint sessions with a biomedical expert from Northwell Health, New York; the findings reported in the abstract and the supplemental materials stem from these sessions, aligning with manual analyses by experts.

AuxiScope: Handheld Augmented Reality Tablet as an Auxiliary Display for Large-Scale Display Systems
Matthew S. Castellana, Chahat Kalsi, Yoonsang Kim, Saeed Boorboor, Arie E. Kaufman
We present AuxiScope, a novel AR-based system designed to enhance personalized data exploration on large wall displays (LWDs) by integrating handheld tablets as auxiliary visualization interfaces. While LWDs offer expanded visual real estate and intuitive embodied interaction, they pose challenges related to effective interfaces for data exploration and analysis, specifically in multi-user settings. AuxiScope addresses these by overlaying supplementary visualizations onto corresponding LWD content, enabling individualized exploration without interference with the visual data displayed on the LWD. To achieve this, we have designed a geometric alignment pipeline that synchronizes the auxiliary visualizations atop the virtual scene. Specifically, by leveraging AR technology, AuxiScope resolves the tablet physical localization, viewpoint computation, and user interaction translation into the virtual space. Subsequently, based on a client-server architecture, it employs remote rendering and delegates computational tasks to the LWD compute nodes in order to minimize memory load on portable devices. We demonstrate the potential of AuxiScope through multiple AR-based interaction techniques across information and scientific visualization scenarios, for both 2D and 3D contexts.
Explainable XR: Understanding User Behaviors of XR Environments using LLM-assisted Analytics Framework
Yoonsang Kim, Zainab Aamir, Mithilesh Singh, Saeed Boorboor, Klaus Mueller, Arie E. Kaufman
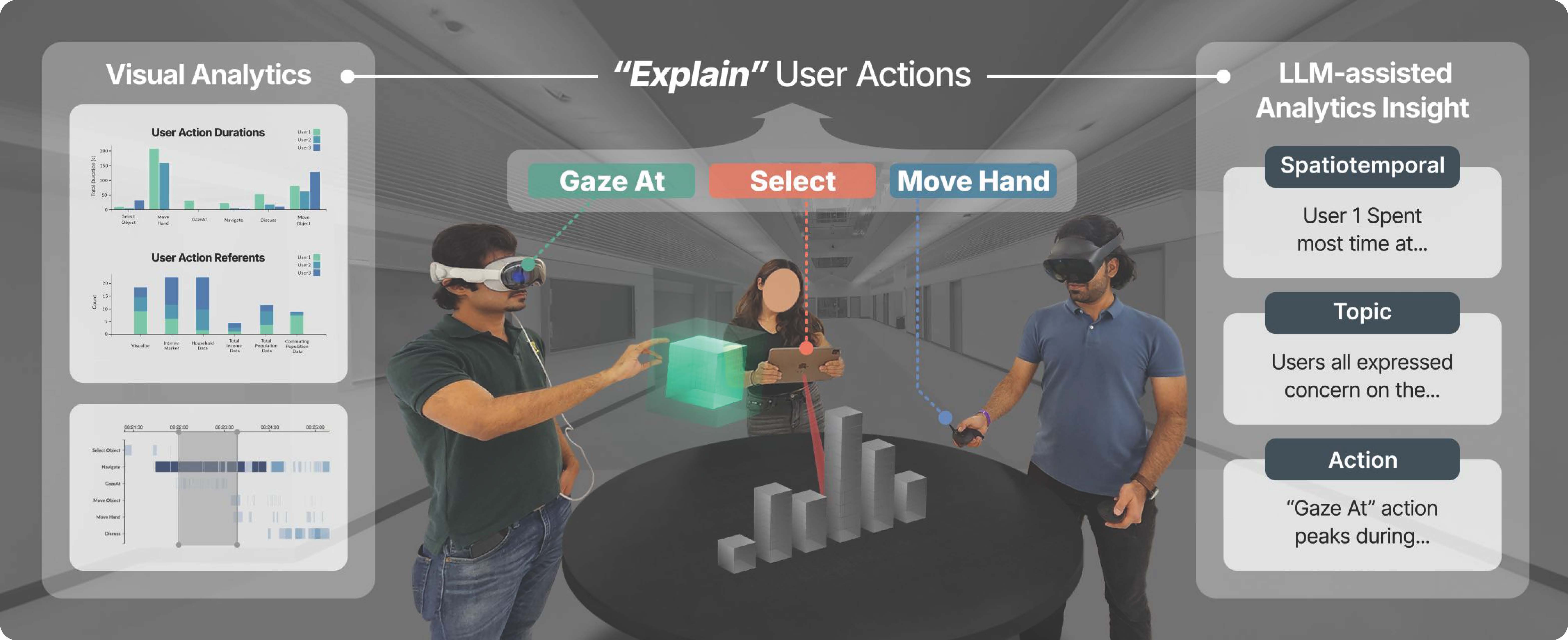
We present Explainable XR, an end-to-end framework for analyzing user behavior in diverse eXtended Reality (XR) environments by leveraging Large Language Models (LLMs) for data interpretation assistance. Existing XR user analytics frameworks face challenges in handling cross-virtuality -- AR, VR, MR -- transitions, multi-user collaborative application scenarios, and the complexity of multimodal data. Explainable XR addresses these challenges by providing a virtuality-agnostic solution for the collection, analysis, and visualization of immersive sessions. We propose three main components in our framework: (1) A novel user data recording schema, called User Action Descriptor (UAD), that can capture the users' multimodal actions, along with their intents and the contexts; (2) a platform-agnostic XR session recorder, and (3) a visual analytics interface that offers LLM-assisted insights tailored to the analysts' perspectives, facilitating the exploration and analysis of the recorded XR session data. We demonstrate the versatility of Explainable XR by demonstrating five use-case scenarios, in both individual and collaborative XR applications across virtualities. Our technical evaluation and user studies show that Explainable XR provides a highly usable analytics solution for understanding user actions and delivering multifaceted, actionable insights into user behaviors in immersive environments.

Silo: Half-Gigapixel Cylindrical Stereoscopic Immersive Display
Saeed Boorboor, Doris E. Gutierrez-Rosales, Ahamed Shoaib, Chahat Kalsi, Yue Wang, Yuyang Cao, Xianfeng Gu, Arie E. Kaufman
We present the design and construction of the Silo, a fully immersive stereoscopic cylindrical tiled-display visualization facility. Comprising 168 high-density LCD displays, the facility provides an ultra-high-resolution image of 619 million pixels, and close to 360 horizontal field-of-regards (FoR), aiming to maximize visual acuity and completely engage the human visual sensorium and its periphery. In this article, we outline the motivations, design principles, hardware selection and software systems, and interaction modalities used in constructing the Silo. To address missing visual information due to the absence of a ceiling and floor, we have designed a method that utilizes conformal mapping and optimal mass transport to reproject the entire 360 volumetric FoR of the virtual scene to the available display real estate. We showcase several applications demonstrating the utility of the Silo and report the findings of our user studies that highlight the effectiveness of the Silo layout compared to curved mono and flat powerwall display facilities. Our user evaluations and studies have shown that the Silo supports natural exploration and enhanced visualization due to its capability to render surround ultra-high-resolution stereoscopic views.
VoxAR: Adaptive Visualization of Volume Rendered Objects in Optical See-Through Augmented Reality
Saeed Boorboor, Matthew S. Castellana, Yoonsang Kim, Chen Zhu-Tian, Johanna Beyer, Hanspeter Pfister, Arie E. Kaufman
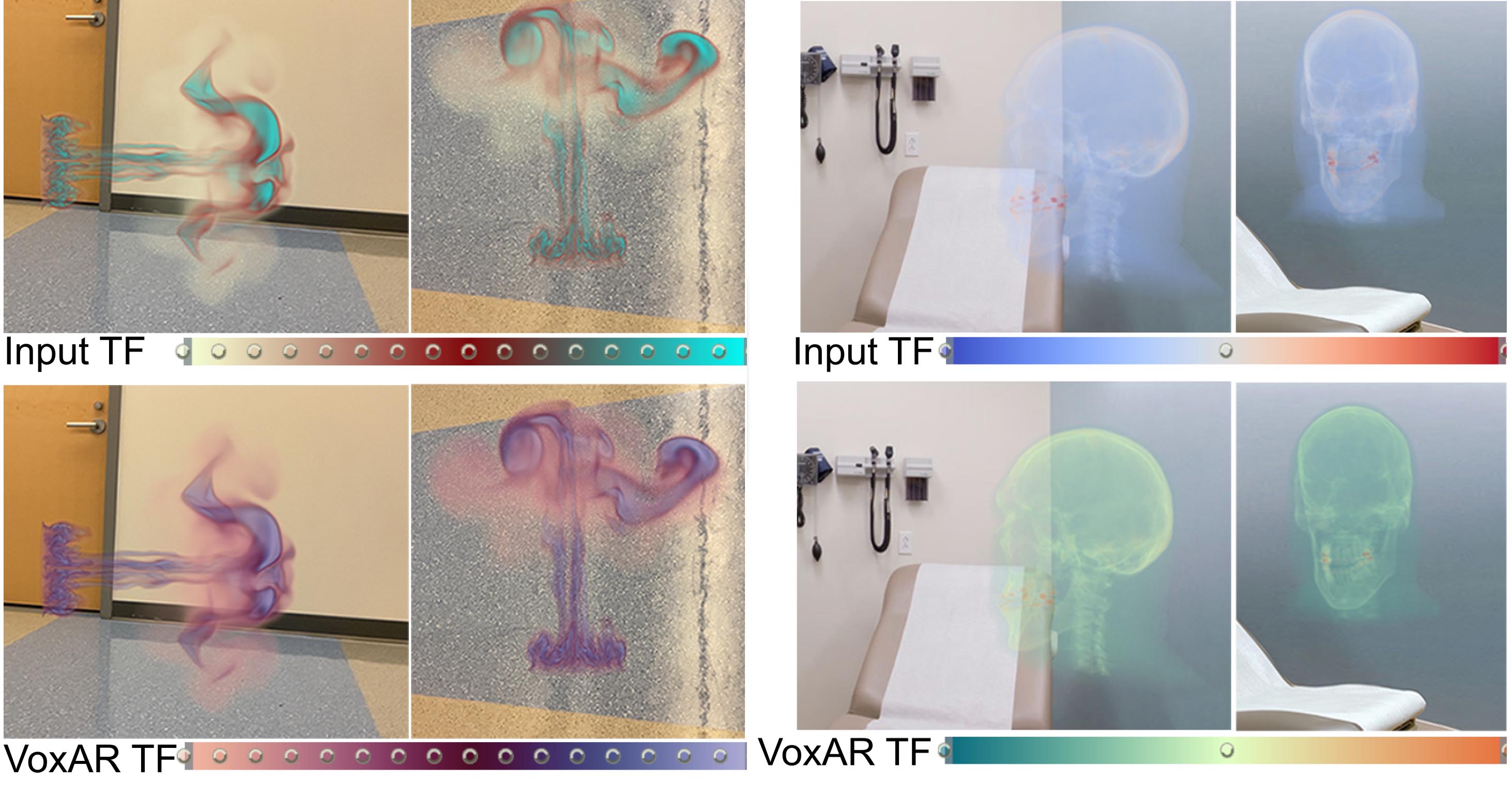
We present VoxAR, a method to facilitate an effective visualization of volume-rendered objects in optical see-through head-mounted displays (OST-HMDs). The potential of augmented reality (AR) to integrate digital information into the physical world provides new opportunities for visualizing and interpreting scientific data. However, a limitation of OST-HMD technology is that rendered pixels of a virtual object can interfere with the colors of the real-world, making it challenging to perceive the augmented virtual information accurately. We address this challenge in a two-step approach. First, VoxAR determines an appropriate placement of the volume-rendered object in the real-world scene by evaluating a set of spatial and environmental objectives, managed as user-selected preferences and pre-defined constraints. We achieve a real-time solution by implementing the objectives using a GPU shader language. Next, VoxAR adjusts the colors of the input transfer function (TF) based on the real-world placement region. Specifically, we introduce a novel optimization method that adjusts the TF colors such that the resulting volume-rendered pixels are discernible against the background and the TF maintains the perceptual mapping between the colors and data intensity values. Finally, we present an assessment of our approach through objective evaluations and subjective user studies.

Submerse: Visualizing Storm Surge Flooding Simulations in Immersive Display Ecologies
Saeed Boorboor, Yoonsang Kim, Ping Hu, Josef Moses, Brian Colle, and Arie E. Kaufman.
We present Submerse , an end-to-end framework for visualizing flooding scenarios on large and immersive display ecologies. Specifically, we reconstruct a surface mesh from input flood simulation data and generate a to-scale 3D virtual scene by incorporating geographical data such as terrain, textures, buildings, and additional scene objects. To optimize computation and memory performance for large simulation datasets, we discretize the data on an adaptive grid using dynamic quadtrees and support level-of-detail based rendering. Moreover, to provide a perception of flooding direction for a time instance, we animate the surface mesh by synthesizing water waves. As interaction is key for effective decision-making and analysis, we introduce two novel techniques for flood visualization in immersive systems: (1) an automatic scene-navigation method using optimal camera viewpoints generated for marked points-of-interest based on the display layout, and (2) an AR-based focus+context technique using an aux display system. Submerse is developed in collaboration between computer scientists and atmospheric scientists. We evaluate the effectiveness of our system and application by conducting workshops with emergency managers, domain experts, and concerned stakeholders in the Stony Brook Reality Deck, an immersive gigapixel facility, to visualize a superstorm flooding scenario in New York City.
NeuRegenerate: A Framework for Visualizing Neurodegeneration
Saeed Boorboor, Shawn Mathew, Mala Ananth, David Talmage, Lorna W. Role, and Arie E. Kaufman.
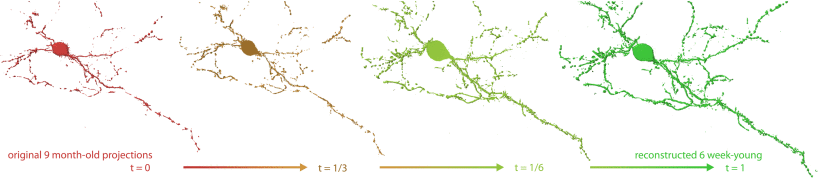
Recent advances in high-resolution microscopy have allowed scientists to better understand the underlying brain connectivity. However, due to the limitation that biological specimens can only be imaged at a single timepoint, studying changes to neural projections over time is limited to observations gathered using population analysis. In this article, we introduce NeuRegenerate , a novel end-to-end framework for the prediction and visualization of changes in neural fiber morphology within a subject across specified age-timepoints. To predict projections, we present neuReGANerator , a deep-learning network based on cycle-consistent generative adversarial network (GAN) that translates features of neuronal structures across age-timepoints for large brain microscopy volumes. We improve the reconstruction quality of the predicted neuronal structures by implementing a density multiplier and a new loss function, called the hallucination loss. Moreover, to alleviate artifacts that occur due to tiling of large input volumes, we introduce a spatial-consistency module in the training pipeline of neuReGANerator. Finally, to visualize the change in projections, predicted using neuReGANerator, NeuRegenerate offers two modes: (i) neuroCompare to simultaneously visualize the difference in the structures of the neuronal projections, from two age domains (using structural view and bounded view), and (ii) neuroMorph , a vesselness-based morphing technique to interactively visualize the transformation of the structures from one age-timepoint to the other. Our framework is designed specifically for volumes acquired using wide-field microscopy. We demonstrate our framework by visualizing the structural changes within the cholinergic system of the mouse brain between a young and old specimen.

Risk Perception and Preparation for Storm Surge Flooding: A Virtual Workshop with Visualization and Stakeholder Interaction
Brian A. Colle, Julia R. Hathaway, Elizabeth J. Bojsza, Josef M. Moses, Shadya J. Sanders, Katherine E. Rowan,
Abigail L. Hils, Elizabeth C. Duesterhoeft, Saeed Boorboor, Arie E. Kaufman, and Susan E. Brennan
Many factors shape public perceptions of extreme weather risk; understanding these factors is important to encourage preparedness. This article describes a novel workshop designed to encourage individual and community decision-making about predicted storm surge flooding. Over 160 U.S. college students participated in this 4-h experience. Distinctive features included 1) two kinds of visualizations, standard weather forecasting graphics versus 3D computer graphics visualization; 2) narrative about a fictitious storm, role-play, and guided discussion of participants’ concerns; and 3) use of an “ethical matrix,” a collective decision-making tool that elicits diverse perspectives based on the lived experiences of diverse stakeholders. Participants experienced a narrative about a hurricane with potential for devastating storm surge flooding on a fictitious coastal college campus. They answered survey questions before, at key points during, and after the narrative, interspersed with forecasts leading to predicted storm landfall. During facilitated breakout groups, participants role-played characters and filled out an ethical matrix. Discussing the matrix encouraged consideration of circumstances impacting evacuation decisions. Participants’ comments suggest several components may have influenced perceptions of personal risk, risks to others, the importance of monitoring weather, and preparing for emergencies. Surprisingly, no differences between the standard forecast graphics versus the immersive, hyperlocal visualizations were detected. Overall, participants’ comments indicate the workshop increased appreciation of others’ evacuation and preparation challenges.
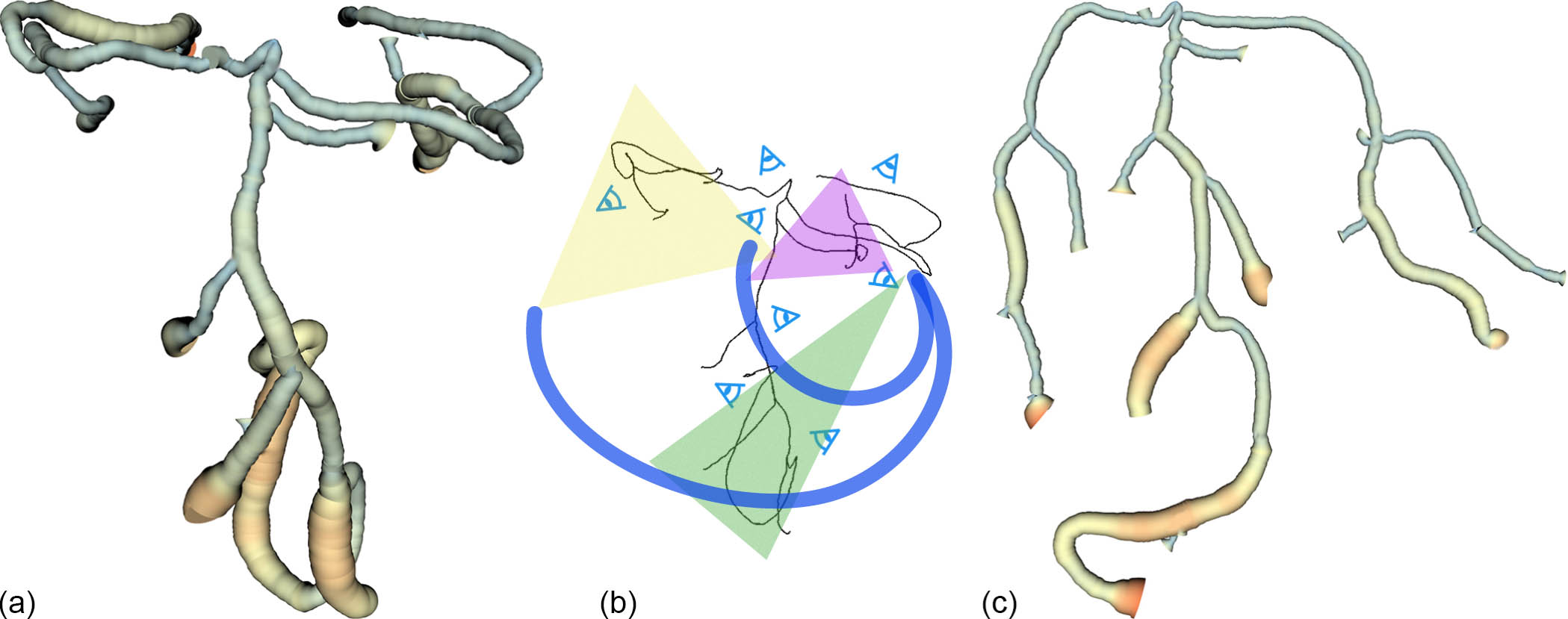
Geometry-Aware Planar Embedding of Treelike Structures
Ping Hu,Saeed Boorboor, Joseph Marino, and Arie E. Kaufman
The growing complexity of spatial and structural information in 3D data makes data inspection and visualization a challenging task. We describe a method to create a planar embedding of 3D treelike structures using their skeleton representations. Our method maintains the original geometry, without overlaps, to the best extent possible, allowing exploration of the topology within a single view. We present a novel camera view generation method which maximizes the visible geometric attributes (segment shape and relative placement between segments). Camera views are created for individual segments and are used to determine local bending angles at each node by projecting them to 2D. The final embedding is generated by minimizing an energy function (the weights of which are user adjustable) based on branch length and the 2D angles, while avoiding intersections. The user can also interactively modify segment placement within the 2D embedding, and the overall embedding will update accordingly. A global to local interactive exploration is provided using hierarchical camera views that are created for subtrees within the structure. We evaluate our method both qualitatively and quantitatively and demonstrate our results by constructing planar visualizations of line data (traced neurons) and volume data (CT vascular and bronchial data).
A Survey of Visualization and Analysis in High-Resolution Connectomics
Johanna Beyer, Jakob Troidl, Saeed Boorboor, Markus Hadwiger, Arie E. Kaufman, and Hanspeter Pfister
The field of connectomics aims to reconstruct the wiring diagram of Neurons and synapses to enable new insights into the workings of the brain. Reconstructing and analyzing the Neuronal connectivity, however, relies on many individual steps, starting from high-resolution data acquisition to automated segmentation, proofreading, interactive data exploration, and circuit analysis. All of these steps have to handle large and complex datasets and rely on or benefit from integrated visualization methods. In this state-of-the-art report, we describe visualization methods that can be applied throughout the connectomics pipeline, from data acquisition to circuit analysis. We first define the different steps of the pipeline and focus on how visualization is currently integrated into these steps. We also survey open science initiatives in connectomics, including usable open-source tools and publicly available datasets. Finally, we discuss open challenges and possible future directions of this exciting research field.
Spatial Perception in Immersive Visualization: A Study and Findings
Ping Hu, Saeed Boorboor, Shreeraj Jadhav, Joseph Marino, Seyedkoosha Mirhosseini, and Arie E. Kaufman
Spatial information understanding is fundamental to visual perception in Metaverse. Beyond the stereoscopic visual cues naturally carried in Metaverse, the human vision system may use other auxiliary information provided by any shadow casting or motion parallax available to perceive the 3D virtual world. However, the combined use of shadows and motion parallax to improve 3D perception have not been fully studied. In particular, when visualizing the combination of volumetric data and associated skeleton models in VR, how to provide the auxiliary visual cues to enhance observers' perception of the structural information is a key yet underexplored topic. This problem is particularly challenging for visualization of data in biomedical research. In this paper, we focus on immersive analytics in neurobiology where the structural information includes the relative position of objects (nuclei / cell body) in the 3D space and the spatial measurement and connectivity of segments (axons and dendrites) in a model. We present a perceptual experiment designed for understanding the consequence of shadow casting and motion parallax in the neuron structures observation and the feedback and analysis of the experiment are reported and discussed.
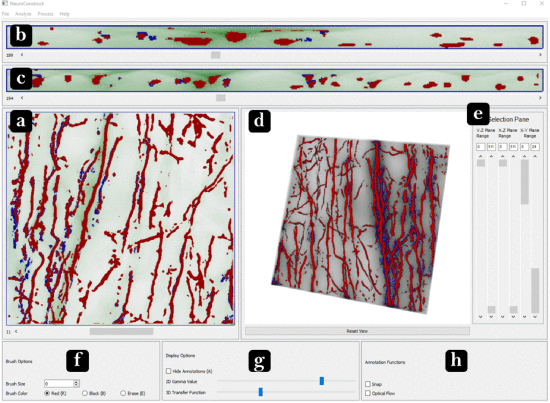
NeuroConstruct: 3D Reconstruction and Visualization of Neurites in Optical Microscopy Brain Images
Parmida Ghahremani, Saeed Boorboor, Pooya Mirhosseini, Chetan Gudisagar, Mala Ananth, David Talmage, Lorna W. Role, and Arie E. Kaufman.
We introduce NeuroConstruct, a novel end-to-end application for the segmentation, registration, and visualization of brain volumes imaged using wide-field microscopy. NeuroConstruct offers a Segmentation Toolbox with various annotation helper functions that aid experts to effectively and precisely annotate micrometer resolution neurites. It also offers an automatic neurites segmentation using convolutional neuronal networks (CNN) trained by the Toolbox annotations and somas segmentation using thresholding. To visualize neurites in a given volume, NeuroConstruct offers a hybrid rendering by combining iso-surface rendering of high-confidence classified neurites, along with real-time rendering of raw volume using a 2D transfer function for voxel classification score versus voxel intensity value. For a complete reconstruction of the 3D neurites, we introduce a Registration Toolbox that provides automatic coarse-to-fine alignment of serially sectioned samples. The quantitative and qualitative analysis show that NeuroConstruct outperforms the state-of-the-art in all design aspects. NeuroConstruct was developed as a collaboration between computer scientists and neuroscientists, with an application to the study of cholinergic neurons, which are severely affected in Alzheimer's disease.
CMed: Crowd Analytics for Medical Imaging Data
Ji Hwan Park, Saad Nadeem, Saeed Boorboor, Joseph Marino, and Arie E. Kaufman.
We present a visual analytics framework, CMed, for exploring medical image data annotations acquired from crowdsourcing. CMed can be used to visualize, classify, and filter crowdsourced clinical data based on a number of different metrics such as detection rate, logged events, and clustering of the annotations. CMed provides several interactive linked visualization components to analyze the crowd annotation results for a particular video and the associated workers. Additionally, all results of an individual worker can be inspected using multiple linked views in our CMed framework. We allow a crowdsourcing application analyst to observe patterns and gather insights into the crowdsourced medical data, helping him/her design future crowdsourcing applications for optimal output from the workers. We demonstrate the efficacy of our framework with two medical crowdsourcing studies: polyp detection in virtual colonoscopy videos and lung nodule detection in CT thin-slab maximum intensity projection videos. We also provide experts’ feedback to show the effectiveness of our framework. Lastly, we share the lessons we learned from our framework with suggestions for integrating our framework into a clinical workflow.
Design of Privacy Preservation System in Augmented Reality
Yoonsang Kim, Saeed Boorboor, Amir Rahmati, and Arie E. Kaufman.
Augmented Reality (AR) capability to overlay virtual data on top of real-world objects and enable better understanding of visual inputs attract the attention of application developers and researchers alike. However, the privacy challenges associated with the use of AR systems is not sufficiently recognized. We present Erebus, a privacy-preserving framework designed for AR applications. Erebus allows the user to establish fine-grained control over the visual data accessible to AR applications. We explore use cases of Erebus framework and how it can be applied to safeguard the privacy of the user’s surroundings in AR environments. We further analyze the latency penalty imposed by Erebus to understand its effect on user experience.
Visualization of Neuronal Structures in Wide-Field Microscopy Brain Images
Saeed Boorboor, Shreeraj Jadhav, Mala Ananth, David Talmage, Lorna Role, and Arie E. Kaufman
Wide-field microscopes are commonly used in neurobiology for experimental studies of brain samples. Available visualization tools are limited to electron, two-photon, and confocal microscopy datasets, and current volume rendering techniques do not yield effective results when used with wide-field data. We present a workflow for the visualization of neuronal structures in wide-field microscopy images of brain samples. We introduce a novel gradient-based distance transform that overcomes the out-of-focus blur caused by the inherent design of wide-field microscopes. This is followed by the extraction of the 3D structure of neurites using a multi-scale curvilinear filter and cell-bodies using a Hessian-based enhancement filter. The response from these filters is then applied as an opacity map to the raw data. Based on the visualization challenges faced by domain experts, our workflow provides multiple rendering modes to enable qualitative analysis of neuronal structures, which includes separation of cell-bodies from neurites and an intensity-based classification of the structures. Additionally, we evaluate our visualization results against both a standard image processing deconvolution technique and a confocal microscopy image of the same specimen. We show that our method is significantly faster and requires less computational resources, while producing high quality visualizations. We deploy our workflow in an immersive gigapixel facility as a paradigm for the processing and visualization of large, high-resolution, wide-field microscopy brain datasets.
Crowdsourcing lung nodules detection and annotation
Saeed Boorboor, Saad Nadeem, Ji Hwan Park, Kevin Baker, and Arie E. Kaufman
We present crowdsourcing as an additional modality to aid radiologists in the diagnosis of lung cancer from clinical chest computed tomography (CT) scans. More specifically, a complete work flow is introduced which can help maximize the sensitivity of lung nodule detection by utilizing the collective intelligence of the crowd. We combine the concept of overlapping thin-slab maximum intensity projections (TS-MIPs) and cine viewing to render short videos that can be outsourced as an annotation task to the crowd. These videos are generated by linearly interpolating overlapping TS-MIPs of CT slices through the depth of each quadrant of a patient's lung. The resultant videos are outsourced to an online community of non-expert users who, after a brief tutorial, annotate suspected nodules in these video segments. Using our crowdsourcing work flow, we achieved a lung nodule detection sensitivity of over 90%for 20 patient CT datasets (containing 178 lung nodules with sizes between 1-30mm), and only 47 false positives from a total of 1021 annotations on nodules of all sizes (96% sensitivity for nodules>4mm). These results show that crowdsourcing can be a robust and scalable modality to aid radiologists in screening for lung cancer, directly or in combination with computer-aided detection (CAD) algorithms. For CAD algorithms, the presented work flow can provide highly accurate training data to overcome the high false-positive rate (per scan) problem. We also provide, for the first time, analysis on nodule size and position which can help improve CAD algorithms.
